Variant Query¶
The authorized users are able to query SNP and INDEL data via gene symbol, rs ID, genomic region, or genomic position. The searching result table includes the information of chromosome name, position, rs ID in dbSNP v149, reference genotype, alteration genotype, variant quality, allele frequency in the ChinaMAP sample sets, gene symbol, affected transcript, and allele frequency in 1000 Genomes database.
Database¶
The following databases are open to query.
ChinaMAP.phase1¶
It contains the information of 136,745,826 SNPs and 10,703,115 INDELs on autosomes from phase 1 study of ChinaMAP. The phase 1 study proposes to analyze 10,588 whole genome sequencing data (~40X) in Chinese natural population.
The query result is shown in the figure below:
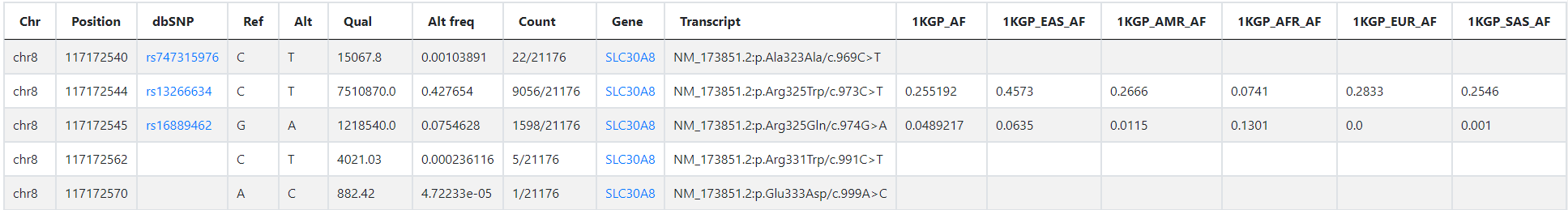
Chr, Chromosome
Position, The reference position
dbSNP, The rs ID in dbSNP
Ref, Reference base
Alt, Alternate base
Qual, Phred-scaled quality score for the assertion made in ALT
Alt freq, Allele Frequency
Count, Allele count in genotypes / Total number of alleles in called genotypes
Gene, Gene symbol
Transcript, Affected transcript
1KGP_AF, Allele Frequency in 1KGP (1000 Genomes Project)
1KGP_EAS_AF, Allele frequency of EAS populations in the 1KGP
1KGP_AMR_AF, Allele frequency of AMR populations in the 1KGP
1KGP_AFR_AF, Allele frequency of AFR populations in the 1KGP
1KGP_EUR_AF, Allele frequency of EUR populations in the 1KGP
1KGP_SAS_AF, Allele frequency of SAS populations in the 1KGP
The flowchart of Variants calling:
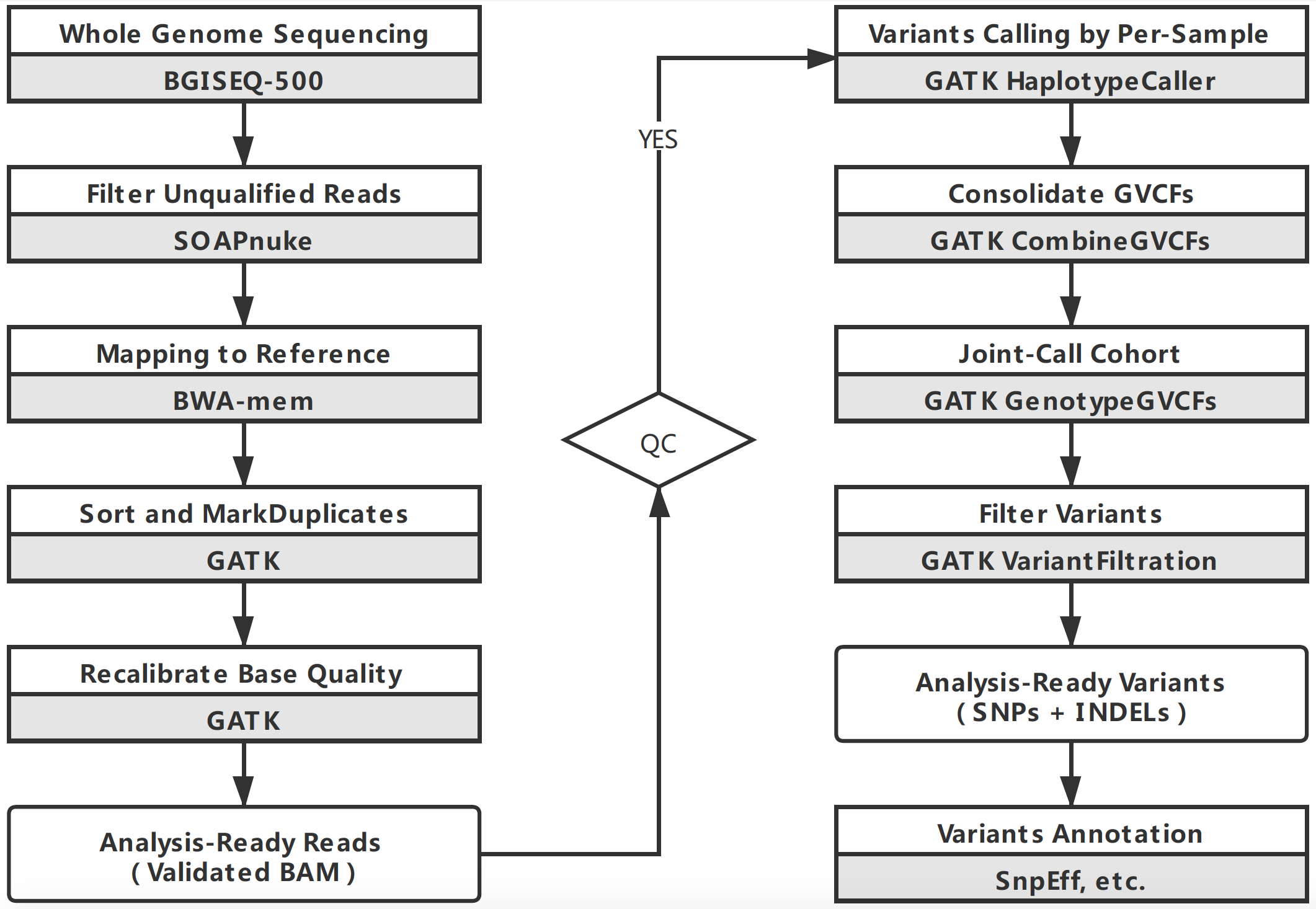
The genome build is GRCh38.